Click Here To Get Powerpoint Presentation
Abstract
Cancer is the second leading cause of death worldwide, following heart diseases. World Health Organization has estimated 12 million deaths worldwide due to cancer in 2030 [1]. According to the American Cancer Society, about 1.5 million new cancer cases and more than 500,000 deaths are expected to occur due to cancer in USA alone in 2009 [2]. Although progress has been steadily made in cancer research to reduce mortality and improve survival, cancer still accounts for nearly 1 in every 4 deaths in the USA [2]. There are many different types of cancer, but all share a common feature – rapid and uncontrolled cell proliferation. Emergence of new tumors is associated with mutations or abnormalities in expression of various cell cycle regulators like cyclins and cyclin-dependent kinases (Cdk) [3]. Therefore, targeting Cdk/cyclin complexes represent a promising therapeutic approach in oncology.
Cell Division Cycle 25 (Cdc25) Phosphatases are Dual-Specificity Phosphatases (DSP) that dephosphorylate and activate the Cdk/cyclin complexes, thereby playing a fundamental role in transitions between cell cycle phases. Also in the event of DNA damage, the Checkpoint Response targets the Cdc25 phosphatases to ensure genomic stability [4,5]. In mammalian cells, three isoforms of Cdc25 phosphatases have been identified : Cdc25A, Cdc25B and Cdc25C. Overexpression of Cdc25A and Cdc25B has been reported in many diverse cancers, thus making them interesting targets for the development of novel anticancer agents [5].
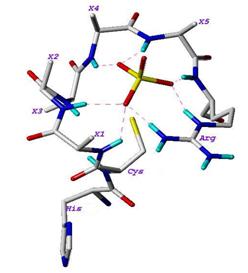
The structure of Cdc25 proteins can be divided into two main regions: the N-terminal region, which is highly divergent and contains sites for its phosphorylation and ubiquitination which regulate the phosphatase activity; and the C-terminal region, which is highly homologous and contains the catalytic site [6]. The crystal structures of the catalytic domains of Cdc25A (PDB ID: 1c25) and Cdc25B (PDB ID: 1qb0) have been solved by X-ray crystallography [7,8]. As shown in Figure 1, the active site loop of Cdc25B has the HCX5R motif, which is highly conserved among the Protein Tyrosine Phosphatase family. H is a highly conserved histidine, C is the catalytic cysteine, and the five Xresidues contribute their backbone amides along with the side chain of arginine (R) to form H-bonds (shown as dashed lines in the figure) with the bound sulfate [8].
A large cavity adjacent to the active site of Cdc25B contains a large number of well-ordered water molecules, called the “swimming-pool” by Rudolph, has shown to accommodate most of the Cdc25B inhibitors [9]. The hotspot residues, which are about 20-30 Å from the active site, are the key elements for substrate recognition [10].
Cdc25 phosphatases are cysteine phosphatases, which contain two important catalytic residues – the catalytic cysteine and a catalytic acid. The cysteine exists as thiolate group at physiological pH with pKa of 5.9 [11]. The thiolate attacks the phosphate of the substrate, followed by protonation of leaving group by the acid and formation of a covalent phospho-cysteine intermediate. The identity of the catalytic acid for Cdc25 phosphatases still remains elusive [11].
A large number of potent small-molecule Cdc25 Inhibitors have been identified that bind to the active site and belong to various chemical classes including natural products, lipophilic acids, quinonoids, electrophilies, sulfonylated aminothiazoles and phosphate bioisosteres [12]. Quinones as inhibitors of Cdc25B will be emphasized during the course of the seminar. The reported quinones that inhibit Cdc25B with IC50 values in the micromolar range (Figure 2) either have a naphthoquinone, quinolinedione, indolyldihydroxyquinone or benzothiazole/benzoxazole-dione core structure [13,14].
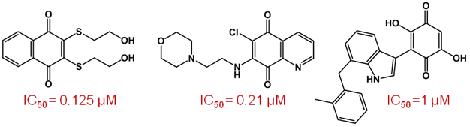
Quinones are electrophilic in nature, suggesting that they might induce a sulfhydryl arylation of cysteine in the catalytic site. They can also inactivate the enzyme by oxidizing the catalytic thiolate group of cysteine. Quinolinediones have displayed mixed inhibition kinetics showing that they can inhibit the enzyme in both reversible and irreversible manners [15]. Molecular modeling studies of these compounds suggested that two different binding modes are possible, one causing reversible inhibition and the other leading to irreversible inhibition, consistent with the mixed inhibition data [16]. From the binding modes of various reversible and irreversible inhibitors of Cdc25B, Lavecchia et al. hypothesized a pharmacophoric model showing the essential structural components for Cdc25B reversible inhibition [17].
Although some progress has been made in developing potent and selective inhibitors for Cdc25 family of proteins, there is scope for development of novel therapeutic strategies to target them. A new class of peptide-derived inhibitors, based on sequence homology with the protein substrate, can be developed. It is challenging to use these compounds as drugs due to their lack of suitable ADME properties [12]. Also, the existence of a potential binding pocket for small molecules adjacent to hotspots on Cdc25B has been proposed [6]. Binding of suitable ligands into this pocket could engage the hotspot residues and interfere in enzyme/substrate association.
Cdc25 phosphatases represent a good target for development of novel antitumor drugs. Crystal structures of Cdc25A and Cdc25B provide a rational basis for the design of potent and selective inhibitors. Further improvement of these inhibitory compounds is likely to lead to their introduction in human clinical trials.
References
- World Health Organization Webpage. http://www.who.int/mediacentre/factsheets/fs297/en/ index.html (accessed 10/22/2009).
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M. J. Cancer statistics, 2009. CA Cancer J Clin. 2009,59, 225-249.
- Johansson, M.; Persson, J. L. Cancer Therapy: Targeting cell cycle regulators. Anti-Cancer Agents Med. Chem. 2008, 8, 723-731.
- Aressy, B.; Ducommun, B. Cell cycle control by the CDC25 phosphatases. Anti-Cancer Agents Med. Chem.2008, 8, 818-824.
- Boutros, R.; Lobjois, V.; Ducommun, B. CDC25 phosphatases in cancer cells: key players? Good targets?Nat. Rev. Cancer. 2007, 7, 495-507.
- Rudolph, J. Cdc25 phosphatases: structure, specificity and mechanism. Biochemistry. 2007, 46(12), 3595-3604.
- Fauman, E. B.; Cogswell, J. P.; Lovejoy, B.; Rocque, W. J.; Holmes, W.; Montana, V. G.; Piwnica-Worms, H.; Rink, M. J.; Saper, M. A. Crystal structure of the catalytic domain of human cell cycle control phosphatase, Cdc25A. Cell. 1998, 93, 617-625.
- Reynolds, R. A.; Yem, A. W.; Wolfe, C. L.; Deibel Jr, M. R.; Chidester, C. G.; Watenpaugh, K. D. Crystal structure of the catalytic subunit of Cdc25B required for G2/M transition of the cell cycle. J. Mol. Biol. 1999,293, 559-568.
- Rudolph, J. Targeting the neighbor’s pool. Mol. Pharmacol. 2004, 66, 780-782.
- Sohn, J.; Kristjansdottir, K.; Safi, A.; Parker, B.; Kiburz, B.; Rudolph, J. Remote hot spots mediate protein substrate recognition for the Cdc25 phosphatases. Proc. Natl. Acad. Sci. USA. 2004, 101, 16437-16441.
- Rudolph, J. Catalytic Mechanism of Cdc25. Biochemistry. 2002, 41, 14613-14623.
- Lazo, J. S.; Wipf, P. Is Cdc25 a druggable target? Anti-Cancer Agents Med. Chem. 2008, 8, 837-842.
- Garuti, L.; Roberti, M.; Pizzirani,D. Synthetic small molecule Cdc25 phosphatase inhibitors. Curr. Med. Chem.2008, 15, 573-580.
- Contour-Galcera, M.; Sidhu, A.; Prevost, G.; Bigg, D.; Ducommun, B. What’s new on Cdc25 phosphatase inhibitors. Pharmacol. Ther. 2007, 115, 1-12.
- Lazo, J. S.; Nemoto, K.; Pestell, K. E.; Cooley, K.; Southwick, E. C.; Mitchell, D. A.; Furey, W.; Gussio, R.; Zaharevitz, D. W.; Joo, B.; Wipf, P. Identification of a potent and selective pharmacophore for Cdc25 dual specificity phosphatase inhibitors. Mol. Pharmacol. 2002, 61, 720-728.
- Lavecchia, A.; Cosconati, S.; Limongelli, V.; Novellino, E. Modeling of Cdc25B dual specificity protein phosphatase inhibitors: docking of ligands and enzymatic inhibition mechanism. ChemMedChem. 2006, 1,540-550.
- Lavecchia, A.; Coluccia, A.; Giovanni, C. D.; Novellino, E. Cdc25B phosphatase inhibitors in cancer therapy: latest developments, trends and medicinal chemistry perspective. Anti-Cancer Agents Med. Chem. 2008, 8, 843-856.

I love to read your articles