DNA transformation is an interesting phenomenon by which one microorganism can transfer genetic material to another microorganism. Transformation occurs by the uptake of exogenous DNA through the cell membrane of the microorganism, followed by its incorporation into the cell as well as expression of the gene. DNA transformation occurs commonly in bacteria, however, it may done with other organisms as well through artificial means. A bacteria that is capable of taking up genetic material from exogenous source is called competent. Artificial transformation makes use of restriction enzymes and DNA ligase. DNA transformation allows the expression of DNA from different species within bacteria, in order to harvest proteins. The following set of two animations goes into the details of how DNA transformation is performed and used.
The animation was provided by the DNA Learning Center and their YouTube Channel. “The mission of the DNA Learning Center is to prepare students and families to thrive in the gene age.”
Animation 1
[swfobject]1476[/swfobject]
Transcript 1
Stanley Cohen and Herbert Boyer’s historic experiment used techniques to cut and paste DNA to create the first custom-made organism containing recombined or “recombinant” DNA. Cohen and Boyer inserted the recombinant DNA molecule they created into E. coli bacteria by means of a plasmid, thereby inducing the uptake and expression of a foreign DNA sequence known as “transformation.”
Stanley Cohen and Herbert Boyer worked together to recombine genes from different bacteria into one DNA molecule. They used genes from two drug-resistant strains of E.coli bacteria- one gene provides resistance to t he antibiotic tetracycline, and the other provides resistance to kanamycin.
Each gene is carried on a plasmid in E. coli. Plasmids are small rings of DNA that exist independently of the main bacterial chromosome. They can be replicated and passed on to offspring.
They named these plasmids p for plasmid and Sc for Stanley Cohen. The plasmid pSC101 carries a gene for tetracycline resistance, and pSC102 carries a gene for kanamycin resistance.
Cohen and Boyer grew the bacterial strains that carried these plasmids, and then isolated the plasmid DNA.
They added the restriction enzyme EcoRI to the plasmid DNA. EcoRI cuts each DNA strand off-center of the recognition site, producing short, single-stranded sequences called “sticky” ends.
They mixed the cut plasmids and added DNA ligase.
Fragments with EcoRI ends are complimentary. This allows fragments to recombine with any other. Hydrogen bonds align two sticky ends, until the ligase repairs the sugar-phosphate bonds to create a stable recombinant molecule.
Before Boyer and Cohen could isolate the recombinant plasmid they wanted, they needed a way to get their ligated plasmids into E.coli. Classic experiments by Oswald Avery and his group showed that Pneumococcus bacteria are “transformed” to virulence when they take up DNA from virulent strains.
However, natural transformation is a rare event, so they used chemical method developed in 1970 by Mandel and Higa at the University of Hawaii. This involved mixing the bacteria and DNA in a suspension of cold calcium chloride at freezing temperature. Then, Boyer and Cohen rapidly raised and lowered the temperature to create a “heat shock”.
This technique induces the bacteria to take in plasmid DNA. They spread the transformed bacteria onto a culture plate containing the antibiotics tetracycline and kanamycin. Only transformed bacteria containing both kinds of resistance genes could grow in the presence of both antibiotics.
This result was consistent with the bacteria being transformed with a recombined plasmid containing both the tetr and the kanr gene.
However, it was also possible that some bacteria had been doubly transformed by relegated versions of the original plasmids.
Restriction analysis showed that some of the colonies had, indeed, been transformed by a recombinant plasmid. Boyer and Cohen were able to tell which was which when they cut the plasmids and ran them out on an agarose gel. (Roll over each band to see the difference.)
Boyer and Cohen had made the first recombinant plasmid. Several months later they showed that these same methods could be used to recombine genes from eukaryotic and prokaryotic organisms. They inserted a frog gene into an E.coli plasmid. The resulting bacteria produced frog DNA.
Animation 2
[swfobject]1477[/swfobject]
Transcript 2
DNA transformation is a naturally occuring but rare event in which DNA can be transferred into bacteria. In 1970, Morton Mandel and Akiko Higa discovered a way to make E. coli more “competent” for transforming foreign DNA. Their calcium chloride method is widely used today to obtain high-efficiency transforming cells.
DNA transformation is a process by which DNA can be transferred into the bacteria. During rapid growth, the cell membrane of E.coli has hundreds of pores, called adhesion zones.
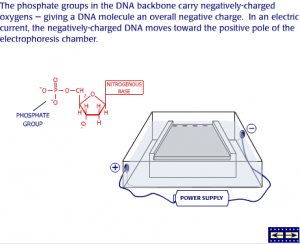
The cell membrane itself is made up of lipid molecules that have negatively-charged phosphates.
Even though the adhesion zones are physically large enough to admit small loops of DNA, the negatively-charged phosphates on the DNA helix are repelled by those on the lipids.
Researchers use a combination of factors to make a bacterial cell capable of taking up new DNA. Theoretically, calcium (Ca2+) ions from added calcium chloride can interact with the negative charges, creating an electrostatically neutral situation.
Lowering the temperature congeals the lipid membrane- stabilizing the negatively-charged phosphates and making them easier to shield.
A rapid rise in temperature, or “heat shock,” then creates a temperature imbalance on either side of the bacterial membrane, and sets up a current. With the “ionic shield” in place, the DNA can then be swept through the adhesion zone.