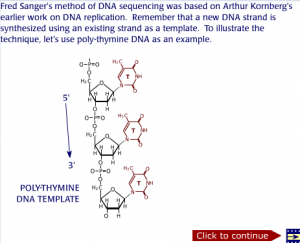
In all cells, the DNA of the cell is responsible for coding the protein sequences. Proteins carry out vital functions in our body. Hence in order to study proteins, their responsible genes encoded in the DNA have to be studied as well. Polymerase chain reaction (PCR) has made it possible for scientists to study many genes by creating multiple copies of the DNA. It allows scientists to make copies of DNA which can then be used to study how different protein and genetic systems work. The advantage of PCR is that it can perform the DNA replication in invitro conditions with the help of thermostable DNA polymerase and heating and cooling. Since the replication of DNA in pcr is invitro, the cycles can be repeated a number of times and in a quick and easy manner to create multiple copies of a single DNA sequence. The animation below demonstrates how Polymerase Chain Reaction (PCR) can be performed and shows how multiple cycles of the polymerase chain reaction can increase the DNA sequence exponentially.
The animation was provided by the DNA Learning Center and their YouTube Channel. “The mission of the DNA Learning Center is to prepare students and families to thrive in the gene age.”
Animation
Transcript
Polymerase chain reaction (PCR) enables researchers to produce millions of copies of a specific DNA sequence in one to two hours. This automated process bypasses the need to use bacteria for amplifying DNA.
PCR: Cycle One
A PCR reaction starts with a denaturing step. Samples are heated to 94-96 °C for 30 seconds or more to denature (separate into single strands) the target DNA.
Next the temperature is lowered to 50-65 °C for thirty seconds or more. This allows the left and the right primers to anneal (base pair) to their complementary sequences. The primers are designed to bracket the DNA region to be amplified.
The temperature is raised to 72 °C for thirty seconds or more. This allows the Taq polymerase to bind at each primed site and extend (synthesize) a new DNA strand.
PCR: Cycle Two
The temperature is raised to 94-96 °C, denaturing the target DNA as in cycle one.
As before, the temperature cools to 50-65 °C allowing more primers to anneal.
The temperature is again raised to 72 °C. Taq polymerase binds to each primed site and synthesizes a new DNA strand.
PCR: Cycle Three
In subsequent cycles, the process of denaturing, annealing and extending are repeated to make additional DNA copies.
PCR: Cycle Four
After three cycles, the target sequence defined by the primers begins to accumulate.
After 30 cycles, as many as billion copies of the target sequence are produced from a single starting molecule.